VOLUME 12 (Supplement)
Philipp. Sci. Lett. 2019 12 (Supplement) 091-100
available online: July 17, 2019
*Corresponding author
Email Address: fe.espino2019@gmail.com
Date Received: February 15, 2019
Date Revised: June 19, 2019
Date Accepted: June 27, 2019
ARTICLE
Detection of antimalarial drug resistance markers in Plasmodium falciparum samples from CARAGA, Philippines: a retrospective study
Edelwisa Segubre-Mercado3, Ronie J. Calugay4,
Sherwin A. Galit1, June Haidee L. Acuna1,
and Fe Esperanza J. Espino*1
Research Institute of Tropical Medicine, Department of Health,
Filinvest Corporate City, Alabang, Muntinlupa City 1781, Philippines
2Department of Biology, School of Science and Engineering,
Ateneo de Manila University, Loyola Heights, Quezon City 1108, Philippines
3Molecular Biology Laboratory, Laboratory Research Division,
Research Institute of Tropical Medicine, Department of Health,
Filinvest Corporate City, Alabang, Muntinlupa City 1781, Philippines
4Department of Biology, College of Science, University of the Philippines
Governor Pack Road, Baguio City 2600, Philippines
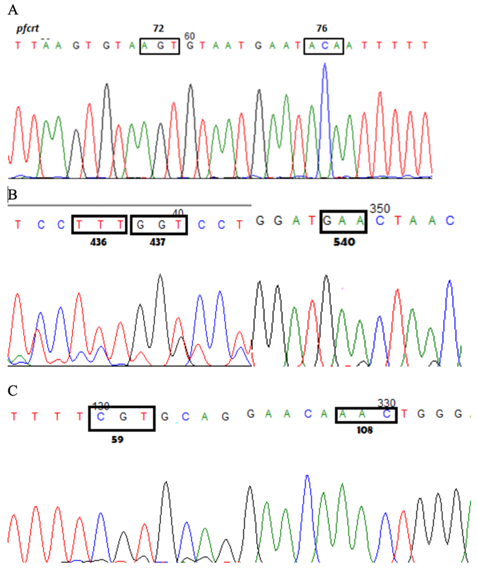
The eradication of malaria globally is hindered by the emergence and spread of antimalarial drug resistance. Therapeutic efficacy surveillance (TES) of antimalarial drugs is done to monitor the susceptibility of malaria parasites to the treatment regimen being implemented. This retrospective study investigated the prevalence of molecular markers associated with resistance to the antimalarial drugs chloroquine (CQ) and sulfadoxine-pyrimethamine (SP) in samples collected from CARAGA (then composed of Agusan del Sur, Agusan del Norte, Surigao del Sur and Surigao del Norte provinces), Mindanao, Philippines from 2005 to 2006 (see Espino et al 2006). DNA from malarial parasites were extracted from 38 dried blood spot samples, and parasite identification in the blood samples by species-specific nested PCR confirmed the cases as Plasmodium falciparum. Single nucleotide polymorphisms in the P. falciparum chloroquine resistance transporter (pfcrt C72S, M74I, N75E/D, and K76T), dihydrofolate reductase (pfdhfr A16V, C50R, N51I, C59R, S108N, and I164L) and dihydropteroate synthetase (pfdhps S436A/F, A437G, K540E, A581G, and A613S/T) genes were determined by nested PCR and sequencing. Genotyping of the msp1, msp2 and glurp polymorphic loci for two samples which had late treatment failure (LTF) identified them as recrudescence. All samples had the pfcrt K76T mutation and harbored the SVMNT mutant haplotype. Moreover, 25% of the isolates were quadruple pfdhps/pfdhfr mutants, with detected polymorphisms at codons 436 and 437 in the pfdhps gene, and codons 59 and 108 in the pfdhfr gene, while 75% were quintuple mutants, including the LTF samples, the additional mutation being codon 540 in the pfdhps gene. No significant difference in the mutation profile was found between recrudescent isolates and those that responded to treatment. These results suggest that mutations that can confer resistance to the administered antimalarial drugs had alarmingly high prevalence in the region. Although previous studies determined the presence of these mutations to be indicative of resistant phenotype, this was not the case for the CARAGA isolates, as samples which harbored mutations still had adequate clinical response. It should be considered that aside from the presence of markers, phenotypic resistance depends on many factors such as host immunity and initial parasite biomass. Nonetheless, the presence of these multiple mutations even sensitive phenotypes could be associated with increased risk of CQ+SP resistance in the CARAGA region thereby providing molecular evidence for the need to shift the national drug policy. The national treatment guidelines were revised in 2009, nevertheless, based on the in vivo results. Further studies can look into the use of these molecular techniques to analyze and compare past and present molecular profiles of antimalarial drug resistance in order to determine possible reversion of resistance. Thus, in vivo studies can be complemented, and a solid basis for formation, execution and validation of public health policies for malaria control and elimination can be provided.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering