VOLUME 16 NUMBER 2 (July to December 2023)
SciEnggJ. 2023 16 (2) 254-264
available online: July 12, 2023
*Corresponding author
Email Address: hbcardos@up.edu.ph
Date received: July 10, 2022
Date revised: May 01, 2023
Date accepted: June 29, 2023
DOI: https://doi.org/10.54645/2023162PUD-98
ARTICLE
Bacterial composition and antibiotic resistome profile of water in the Manila Bay South Harbor
Molecular Biology, College of Medicine, Manila, Philippines
2University of the Philippines Manila, Department of Biology,
College of Arts and Sciences, Manila, Philippines
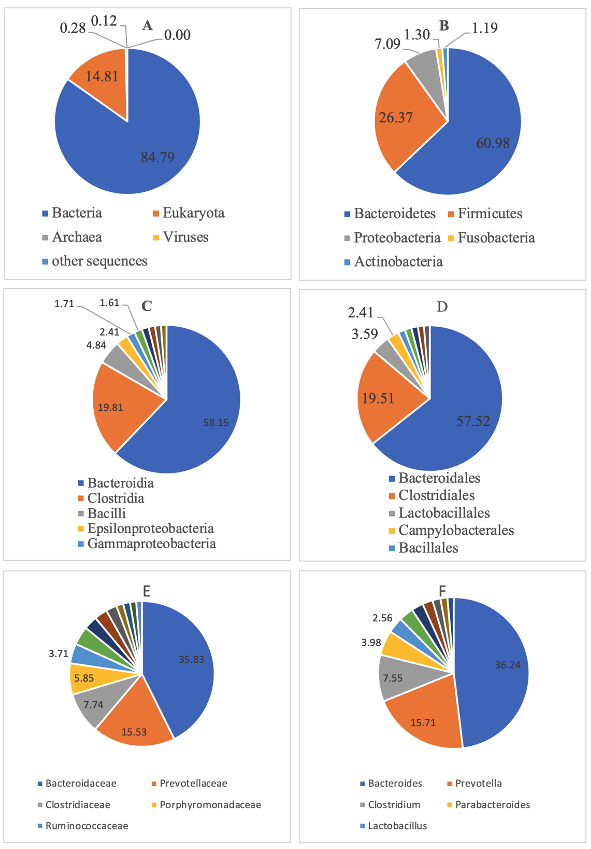
Anthropogenic activities in Manila Bay, the oldest and busiest international port in the Philippines, may cause changes in flora and fauna within its harbors. Specifically, the ballasting and deballasting of ships may accidentally introduce new populations of microorganisms, plants, and animals. Ballasting (and deballasting), or the process of carrying in/out of seawater from ship tanks in major ports/harbors, are causing major disturbances, particularly in marine ecosystems. Previous studies have shown that marine microorganisms comprise 98% of the primary production and regulation of the ocean's biogeochemical processes. However, with the continuous community disturbances due to human influence, potential marine bacterial populations with antibiotic resistomes may occur, and impact marine diversity. Since data on the microbial communities in Manila Bay South Harbor are limited, this study determined the bacterial composition and antibiotic resistome profiles of water samples from the area. Next-generation sequencing techniques and culture-based methods were utilized to elucidate the bacterial and functional diversity, as well as the antibiotic resistome of the water samples from the Manila Bay South Harbor. The bacterial diversity of the water sample was made up primarily of members of Bacteroidetes, Firmicutes, and Prevotella. The Clusters of Orthologous Group (COG) Level 1 identified metabolism as the most dominant functional predicted metagenome. The characterization of antibiotic resistance genes (ARGs) from water samples revealed that ARGs encoding resistance to multidrug was the most abundant. On the other hand, the identified antibiotic-resistant bacteria included ampicillin-resistant Escherichia coli, multidrug-resistant Klebsiella pneumoniae, ampicillin- and cefuroxime-resistant Vibrio alginolyticus and ampicillin resistant Vibrio furnissii. The above results provide a baseline marine bacterial community and antibiotic resistome profile of the Manila Bay South Harbor. This information is critical in gaining a better understanding of the potential roles of shifts in marine microbiomes, and in turn, help in developing appropriate solutions for managing the undesirable consequences of ballast water.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering