VOLUME 16 NUMBER 2 (July to December 2023)
SciEnggJ. 2023 16 (2) 442-457
available online: December 12, 2023
*Corresponding authors
Email Addresses: lynnesther.rallos@msugensan.edu.ph; wlrivera@science.upd.edu.ph
Date received: October 15, 2023
Date revised: December 5, 2023
Date accepted: December 6, 2023
DOI: https://doi.org/10.54645/2023162XWH-26
ARTICLE
Profiling Lignocellulose Degraders in the Microbiome of Rice Straw from Three Selected Lowland Farms in General Santos City, Philippines Using a High-Throughput Microarray Platform
Mindanao State University–General Santos, Fatima, General Santos City,
Philippines
2Pathogen–Host–Environment Interactions Research Laboratory, Institute of
Biology, College of Science, University of the Philippines Diliman,
Quezon City, Philippines
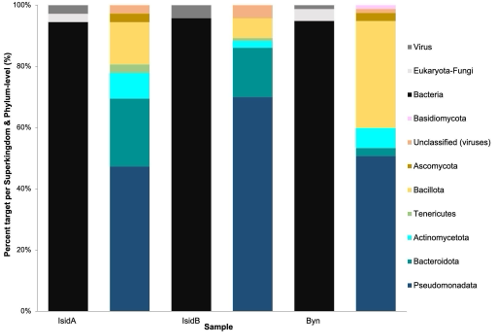
Crop residues, such as rice straw, usually regarded as agricultural wastes recalcitrant to degradation, offer a myriad of services in the field, including reservoirs of valuable microorganisms and bioactive substances that are necessary for productive agroecosystems. However, very few studies report on the utilization of rice straw as a source of lignin-degrading microorganisms. This study probed into the taxonomic diversity of microorganisms associated with rice straw using metagenomic DNA obtained from samples collected in three lowland rice fields in General Santos City, Philippines for bioprospecting of microorganisms with ligninolytic potential and expanding the knowledge on prokaryotic lignin-degraders. Three samples collected from each site were pooled into one sample for the extraction of genomic DNA that was further subjected to the AxiomTM Microbiome Array (AMA) analysis, a relatively new high-throughput detection system, that can comprehensively identify a wide range of microorganisms at the species and strain levels. The sequences identified via AMA analysis were distributed across 3 superkingdoms, 7 phyla and 55 families. Our data showed that the samples contain a few fungi and some DNA viruses but are primarily composed of prokaryotic bacteria, with Actinomycetota, Bacillota, Bacteroidota, and Pseudomonadota being the dominant phyla in terms of the number of detected species. The rice straw microbiomes harbored several microbial species that have been widely reported elsewhere as lignocellulose degraders. Several noteworthy species detected by AMA were closely related to Bacteroides thetaiotaomicron, Cellvibrio japonicus, Pseudomonas putida, Flavobacterium rivuli, Fusarium graminearum, and Streptomyces sp. The results of this study provide molecular evidence that rice straw-associated microbiomes are rich sources of biological agents for crop residue management.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering