VOLUME 16 (Supplement)
SciEnggJ 16 (Supplement) 107-129
available online: November 28, 2023
DOI: https://doi.org/10.54645/XSNN78625
*Corresponding author
Email Address: aolluisma@up.edu.ph
Date received: September 10, 2023
Date revised: October 27, 2023
Date accepted: October 31, 2023
DOI: https://doi.org/10.54645/XSNN78625
ARTICLE
Genome Mining of a Novel Marine Sponge Symbiont Nocardia sp. BML-15-R-026U Reveals High Biosynthetic Potential for Secondary Metabolites, Including a Non-Ribosomal Peptide and a Polyketide of High Novelty
Quezon City
2Institute of Biology, University of the Philippines, Diliman, Quezon City
3Marine Science Institute, University of the Philippines, Diliman,
Quezon City
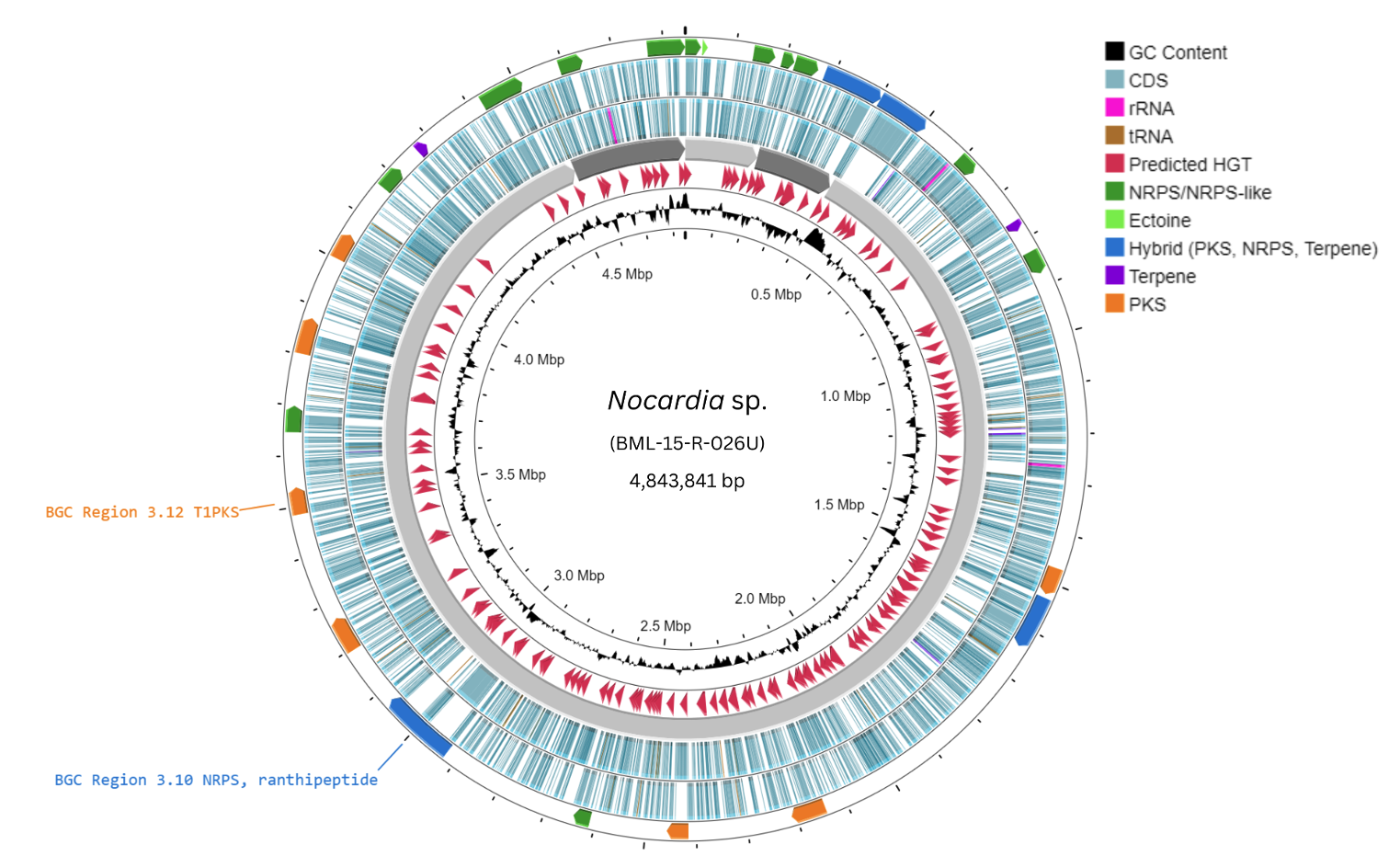
Antibiotic and drug resistance poses serious global public health threats, leading to substantial infections and fatalities annually. Addressing these issues requires the discovery of novel bioactive compounds and a faster and more cost-effective discovery process. However, traditional approaches, which require isolation and multi-step purification of compounds from organisms and running of initial assays, suffer from serious limitations such as the need for substantial amounts of biological material and high rates of compound rediscoveries. Because the biosynthetic capabilities of organisms are encoded in their genomes, genome mining provides a promising solution that would complement traditional approaches. This study conducted long-read whole genome sequencing on a marine sponge symbiont, Nocardia sp. BML-15-R-026U, to explore its genomic repertoire of secondary metabolite-encoding Biosynthetic Gene Clusters (BGCs). A four-contig genome assembly was generated for this isolate with a high degree of completeness and an estimated genome size of 4.84 Mbp. Its genome displays remarkable biosynthetic potential by containing at least 34 distinct secondary metabolite BGCs, predominantly Non-Ribosomal Peptide Synthetase (NRPS) and Polyketide Synthase (PKS) systems capable of producing novel chemical structures. Further analysis was focused on two genomic regions. In region 3.10, the study predicted a BGC for a novel, serine-rich non-ribosomal peptide with a predicted molecular weight of 2754 g/mol. Region 3.12 contained an iterative type-I PKS BGC, suggesting the potential synthesis of a polyketide compound with oxidoreductase-inhibiting properties. This study highlights genome mining as a productive early-phase approach for identifying promising drug leads and has identified the most promising candidates among this isolate’s BGCs for experimental validation.
© 2025 SciEnggJ
Philippine-American Academy of Science and Engineering