VOLUME 18 NUMBER 1 (January to June 2025)
SciEnggJ. 2025 18 (1) 177-190
available online: 19 May 2025
DOI: https://doi.org/10.54645/2025181XOI-52
*Corresponding author
Email Address: nssantos1@up.edu.ph
Date received: 17 July 2024
Dates revised: 23 September 2024;
02 November 2024; 24 February 2025
Date accepted: 27 April 2025
ARTICLE
Comparative sequence analysis of candidate antifungal defense genes within the SH3 genomic region of Arabica, Robusta, Liberica, and Excelsa coffee grown in the Philippines
Biology, College of Science University of the Philippines
Diliman, Quezon City, 1101
2Science and Technology Scholarship Division, Science Education
Institute, Department of Science and Technology, Bicutan,
Taguig City, 1631
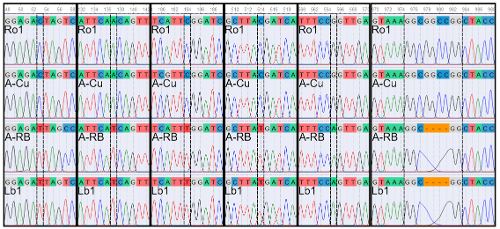
The SH3 genes are a group of well-studied antifungal coffee defense genes, which have been localized to a region in chromosome 3. A previous study screened this region for other antifungal defense genes, particularly, those implicated in defense against coffee leaf rust (CLR) and coffee berry disease (CBD). Three candidate defense gene loci have been identified, namely, an ethylene-responsive transcription factor 1B (ERF1B) gene, a chalcone synthase 2-like (CHS2) gene, and a major allergen Pru ar 1-like gene. For this study, the main goal is to evaluate and compare the sequences of these selected loci from a sample of Philippine coffee accessions, which consist of the four main varieties utilized in local production: Arabica (Coffea arabica), Robusta (C. canephora), Liberica (C. liberica var. liberica) and Excelsa (C. liberica var. dewevrei). DNA was extracted from young leaves and subjected to PCR amplification using newly designed primers. The PCR products were run through agarose gel electrophoresis after which, gel portions containing the target bands were excised and processed for bidirectional sequencing. Consensus sequences were constructed from the raw sequences using UGENE. Due to probable primer specificity issues, the CHS2 sequences were excluded from further analysis. In order to analyze the ERF1B and Pru ar 1-like gene sequences, MEGA11 was used for multiple sequence alignment, protein sequence prediction, and construction of neighbor-joining trees. The sequences were also cross-referenced against the GenBank database using BLAST. Results show that several of the sequences represent novel orthologs, especially those obtained from the Liberica and Excelsa samples. Furthermore, orthologs which are most likely linked to conferring greater resistance were determined. Most notable among these are the ERF1B sequences of a Red Bourbon (A-RB, NSIC-2008-Cf-A-05) and a Mundo Novo (A-MN) sample. Despite originating from Arabica samples, the A-RB and A-MN ERF1B orthologs show greater similarity to their counterparts in Liberica/Excelsa and Robusta samples, respectively. These orthologs are recommended for follow-up studies through additional in silico analysis and genotype-phenotype association so they can definitively be considered as target genes in future coffee breeding programs.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering