VOLUME 18 NUMBER 1 (January to June 2025)
SciEnggJ. 2025 18 (1) 210-228
available online: 22 June 2025
DOI: https://doi.org/10.54645/2025181KIA-75
*Corresponding author
Email Address: abcustodio@up.edu.ph; sasedano@up.edu.ph
Date received: 15 January 2025
Dates revised: 19 April 2025; 28 May 2025
Date accepted: 10 June 2025
ARTICLE
Characterization of attributes beneficial to pathogen fitness of Philippine Streptococcus suis strain 8-57C serotype 1/2 from diseased pig lung
University of the Philippines – Los Baños, College, Laguna
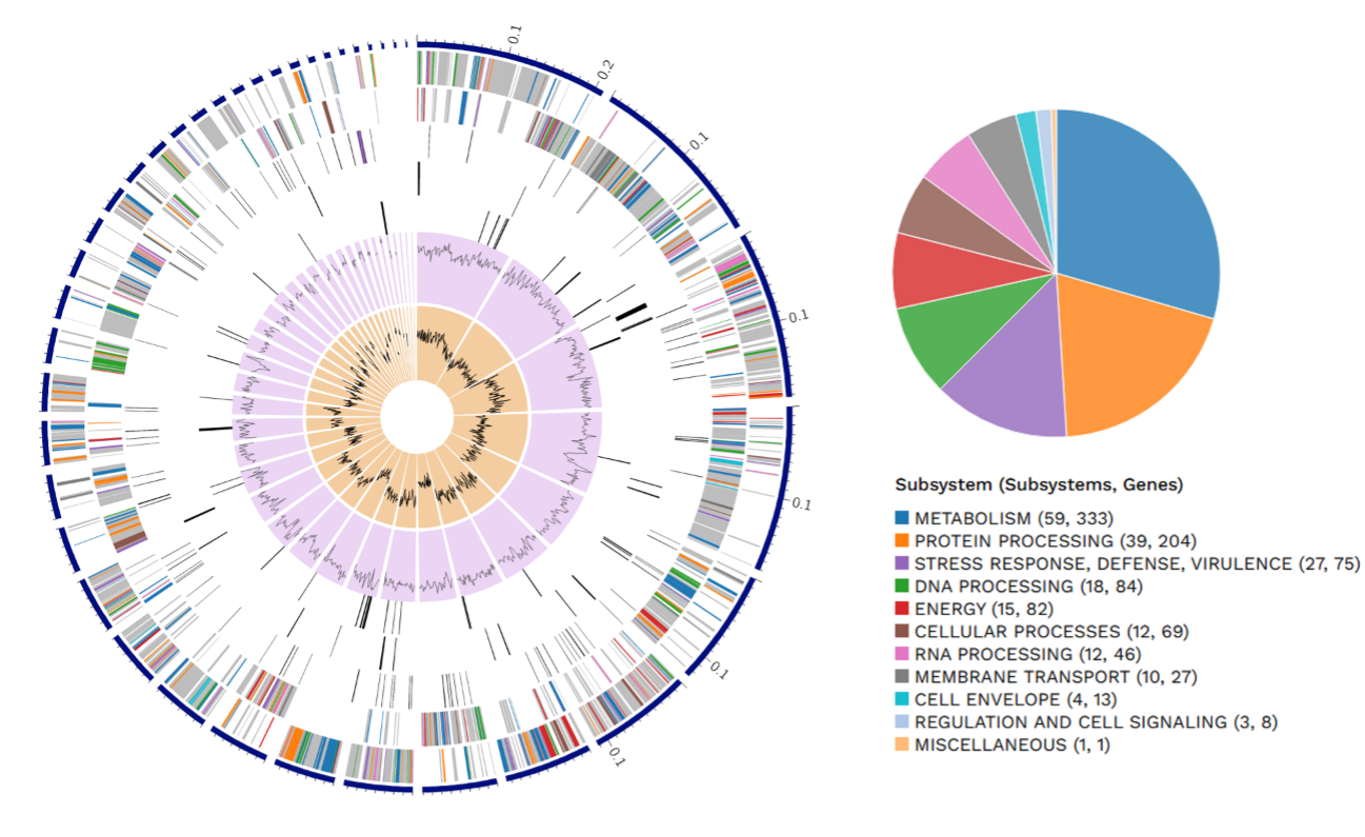
Research on the important swine zoonotic pathogen Streptococcus suis is disproportionately directed towards serotype 2 since this is the most common and virulent serotype. However, mechanisms of epidemiology and pathogenesis of possibly opportunistic serotypes like serotype 1/2, also common in pigs, are usually understudied. S. suis research in swine in the Philippines is severely lacking, with only two publications in the past half-decade. Hence, this study performed a genome-based evaluation of the Philippine S. suis strain 8-57C serotype 1/2 isolated from diseased pig lung to determine characteristics beneficial to pathogen fitness, like the presence of antimicrobial resistance (AMR) genes, virulence factors (VFs), and biosynthetic gene clusters (BGCs). PCR-based AMR gene screening and antimicrobial susceptibility testing were also performed to verify the predicted AMR activity. Genome analysis revealed the presence of several VF classes (adherence, enzyme, immune evasion, and protease) which could aid pathogenesis with the ability to adhere to cells and organs, cross the blood-brain barrier, protect against phagocytosis, and exhibit hemolytic activity. Genome-based AMR prediction, PCR-based AMR profiling, and antimicrobial susceptibility testing showed that S. suis strain 8-57C was resistant to lincosamides, macrolides, streptogramin B, tetracycline, and amphenicol antibiotic classes. Additionally, BGC prediction identified a lanthipeptide BGC similar to the bacteriocin suicin 90-1330. Further experimentation is recommended to verify if these virulence factors and metabolites are expressed and if they contribute to pathogen fitness. These findings could pave the way for exploring characteristics relevant to the pathogen fitness of other non-serotype 2 S. suis serovars in the Philippines.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering