VOLUME 18 NUMBER 2 (July to December 2025)
SciEnggJ. 2025 18 (2) 275-294
available online: 22 August 2025
DOI: https://doi.org/10.54645/2025182HWH-76
*Corresponding author
Email Address: mpdeleon1@up.edu.ph
Date received: 07 April 2025
Dates revised: 10 July 2025
Date accepted: 13 August 2025
ARTICLE
Mapping the bacterial diversity and functional landscape of the Philippine bat gut microbiome using a 16S-based metataxonomics approach
Los Baños, College, Los Baños, Laguna, Philippines
2Environmental Biology Division, Institute of Biological Sciences,
University of the Philippines Los Baños, College, Los Baños,
Laguna, Philippines
3Microbiology Division, Institute of Biological Sciences,
University of the Philippines Los Baños, College, Los Baños,
Laguna, Philippines
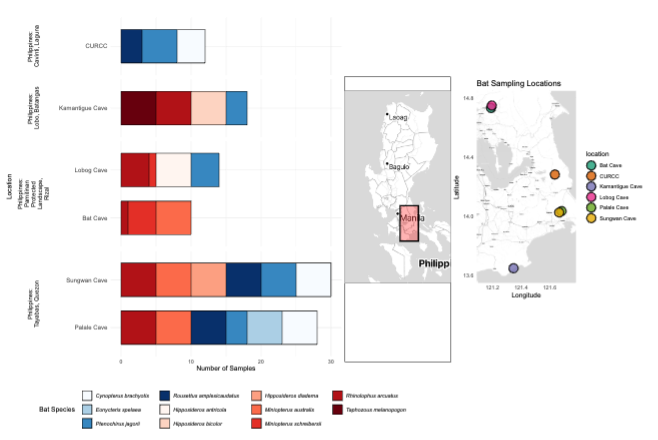
Understanding the gut microbiome of bats is essential for elucidating its role in host health, metabolism, and the transmission of potentially zoonotic pathogens. Despite this, the gut microbiomes of Philippine bats remain largely uncharacterized. In this study, we present a comprehensive examination of the gut microbiome profiles of 112 bats captured from multiple cave sites in the CALABARZON region of Luzon, Philippines. Using 16S ribosomal RNA (rRNA) gene amplicon sequencing, we compared the microbiomes of various insectivorous (n=60) and frugivorous (n=52) bat species, accounting for dietary niches, host identity, and location. In terms of microbial diversity, results reveal that insectivorous bats show significantly lower Shannon diversity (H’) and Inverse Simpson metrics than frugivorous bats (K-W; p<0.05). Beta diversity analysis indicates a strong influence of cave location on microbial community composition, while diet had weaker effects—suggesting overlap and similarity of gut communities across dietary guilds. Stratified permutational analysis also highlights notable convergence among bats within the same dietary guild or species roosting at different caves, particularly among frugivorous bats such as Ptenochirus jagori. Compositional analysis shows a predominance of Pseudomonadota and Bacillota across samples, with several frugivorous bats harboring potentially pathogenic families (Neisseriaceae, Campylobacteraceae, Mycoplasmoidaceae, Helicobacteraceae). Functional predictions using PiCRUSt2 suggest that insectivores, despite their lower taxonomic diversity, possess higher functional diversity, notably in amino acid metabolism pathways. This enrichment may reflect specific metabolic adaptations or dietary requirements. These interesting findings elucidate the role of ecological factors in shaping the gut microbiome of cave bats, particularly in the South Luzon region.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering