VOLUME 18 NUMBER 2 (July to December 2025)
SciEnggJ. 2025 18 (2) 383-395
available online: 27 October 2025
DOI: https://doi.org/10.54645/2025182SFO-23
*Corresponding author
Email Address: lmdalmacio@up.edu.ph
Date received: 01 March 2025
Dates revised: 20 July 2025, 29 September 2025
Date accepted: 06 October 2025
ARTICLE
Wastewater surveillance of SARS-CoV-2 RNA and detection of emerging viral variants from wastewater in the city of Manila, Philippines
Philippines
2Graduate School, University of the Philippines Los Baños,
Los Baños, Laguna, Philippines
3Gokongwei College of Engineering, De La Salle University Manila,
Manila, Philippines
4Graduate School, Angeles University Foundation, Pampanga,
Philippines
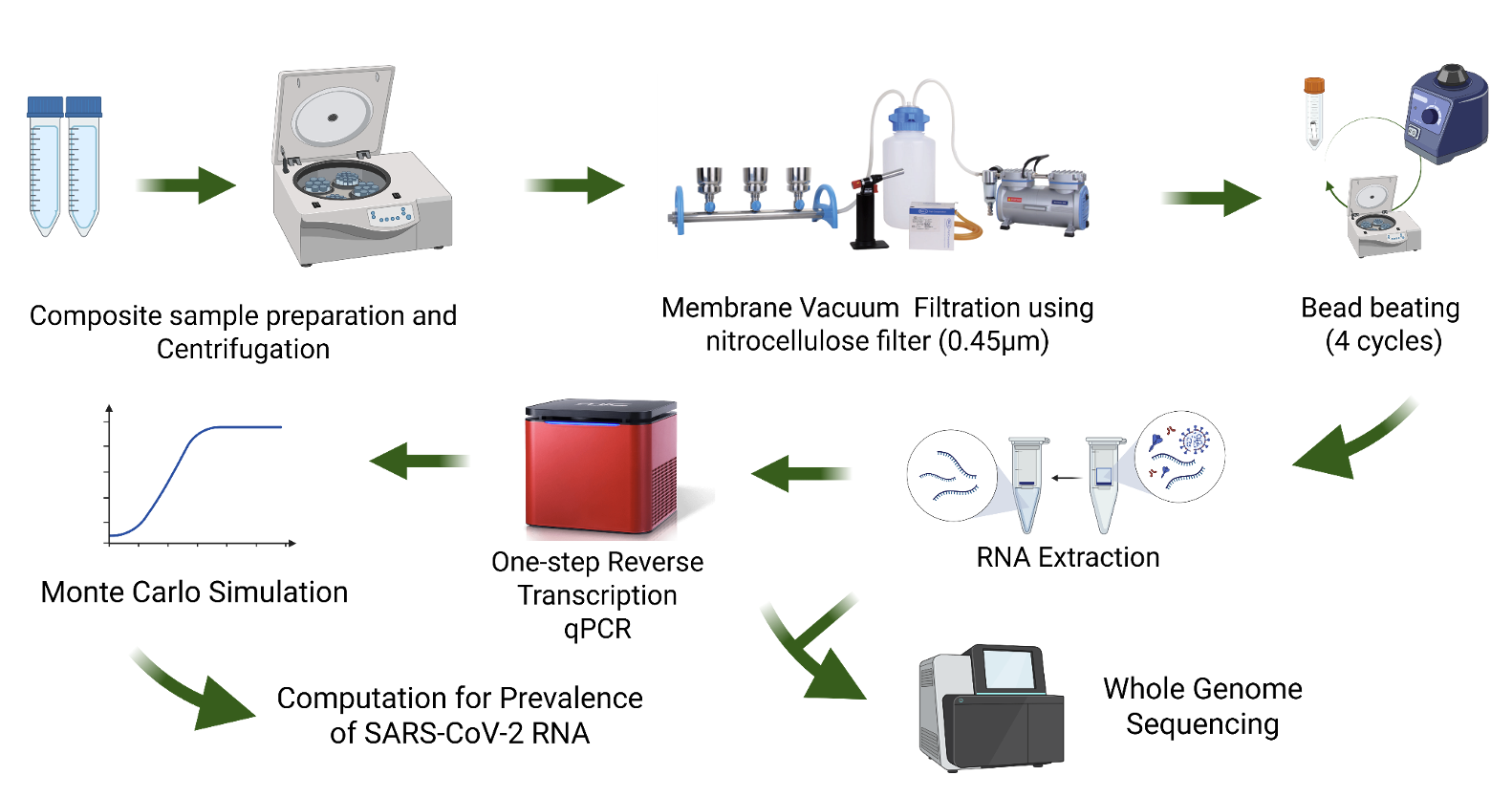
Wastewater-based epidemiology (WBE) of SARS-CoV-2 has been explored in many countries during the past years of the COVID-19 pandemic (2021-2022) along with cost-efficient large-scale monitoring of infection. In this study, samples (n=88) were collected for three consecutive months at four different sites (A - hospital, B - immediate hospital-government offices, C - residential, D - residential-commercial). RT-qPCR was used to quantify SARS-CoV-2 RNA, concentrations, specifically the N1 and N2 genes, in samples collected from a sewer network from a hospital to a community in the City of Manila, Philippines. Monte Carlo Simulation software was then used to compute the viral RNA prevalence in the wastewater matrix. Whole genome sequencing was then conducted to determine the emerging SARS-CoV-2 viral variants in wastewater. Results showed a higher detection rate of N1 genes compared to N2 genes. The highest concentration of viral RNA was obtained from Site A, while the lowest concentration was obtained from Site C. All 13 sequenced samples were representatives of Omicron variants, five of which were detected prior to their first detection in clinical samples. Notably, three Omicron subvariants were detected in both wastewater and clinical samples within the same sampling period. These show that SARS-CoV-2 RNA has potential as a biomarker for WBE in Manila, Philippines. However, further studies are needed to optimize the viral RNA recovery protocols and plans to recommend policies for efficient sewer line management should go hand in hand for the successful integration of this system in the City of Manila.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering