VOLUME 18 (Supplement)
SciEnggJ 18 (Supplement) 493-499
available online: 22 December 2025
DOI: https://doi.org/10.54645/202518SupREA-14
*Corresponding author
Email Address: wlrivera@science.upd.edu.ph
Date received: 22 September 2025
Date revised: 25 November 2025
Date accepted: 11 December 2025
ARTICLE
A comparative study of culture-based and PCR-based protocols for Salmonella enterica detection in meat, environmental swabs, and commercial animal feed
Institute of Biology, College of Science, University of the
Philippines Diliman, Quezon City 1101, Philippines
2Microbiology Section, Laboratory Division, National Meat
Inspection Service, Department of Agriculture, Quezon City
1100, Philippines
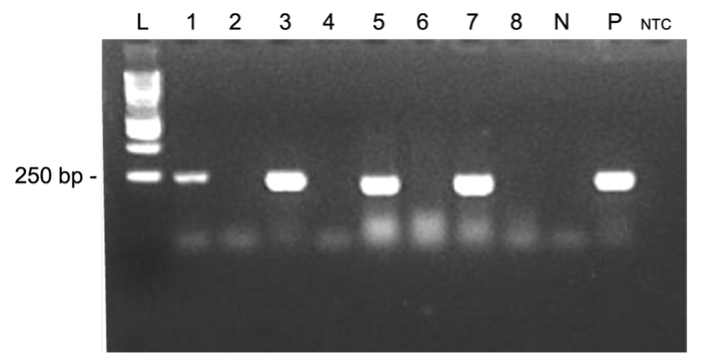
A polymerase chain reaction (PCR)-based assay targeting the Salmonella-specific invA gene was validated for the detection of Salmonella enterica in beef, chicken, animal feed, and sponge swabs. Assessed by a third-party proficiency testing service, the protocol was rated ‘Satisfactory’ across all matrices. It consistently demonstrated effective in detecting S. enterica not only in meat but also in non-meat sources, such as animal feed and environmental swabs. To compare performance with the conventional culture-based method outlined in the Bacteriological Analytical Manual (BAM) and ISO 6579-1:2017(E), parallel testing was conducted by the National Meat Inspection Service (NMIS), Department of Agriculture, Philippines, and the Pathogen-Host-Environment Interactions Research Laboratory (PHEIRL), University of the Philippines Diliman. Results showed that 84 of 95 samples (88%) yielded consistent outcomes between culture and PCR. However, 11 samples (12%) tested negative by culture but positive by PCR, likely due to atypical colonies that lacked characteristic morphology on the culture media. Findings confirm the PCR assay’s superiority for S. enterica detection across diverse food and non-food matrices, regardless of colony morphology. Moreover, the assay provides results within two days, compared to five to seven days required by traditional methods, highlighting its value for rapid diagnostics. Notably, the study also reports the detection of S. enterica in commercial feed, underscoring its potential role as a disease vector. This is timely and significant, as the Philippines currently lacks microbiological quality standards for finished feed products, with testing limited to raw materials. The study provides critical groundwork for establishing microbiological standards for finished feed products in the country.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering