VOLUME 18 (Supplement)
SciEnggJ 18 (Supplement) 515-538
available online: 31 December 2025
DOI: https://doi.org/10.54645/202518SupEOX-42
*Corresponding author
Email Address: cdlaurenciano@ust.edu.ph
Date received: 08 August 2025
Dates revised: 01 October 2025, 31 October 2025,
18 December 2025
Date accepted: 25 December 2025
ARTICLE
Philippine marine natural product scaffolds as inhibitors of DNA damage response pathways with promising binding propensity, stability, and pharmacokinetic profiles
University of Santo Tomas, 1008 Manila, Philippines
2UST Laboratories for Vaccine Science, Molecular Biology, and
Biotechnology, Research Center for Natural and Applied
Sciences, University of Santo Tomas, 1008 Manila, Philippines
3Chemistry Department, College of Science and Mathematics,
Mindanao State University – Iligan Institute of Technology,
Tibanga 9200, Iligan City, Philippines
4Department of Science and Technology - Philippine Science High
School – Central Mindanao Campus, 9217 Balo-i, Lanao del
Norte, Philippines
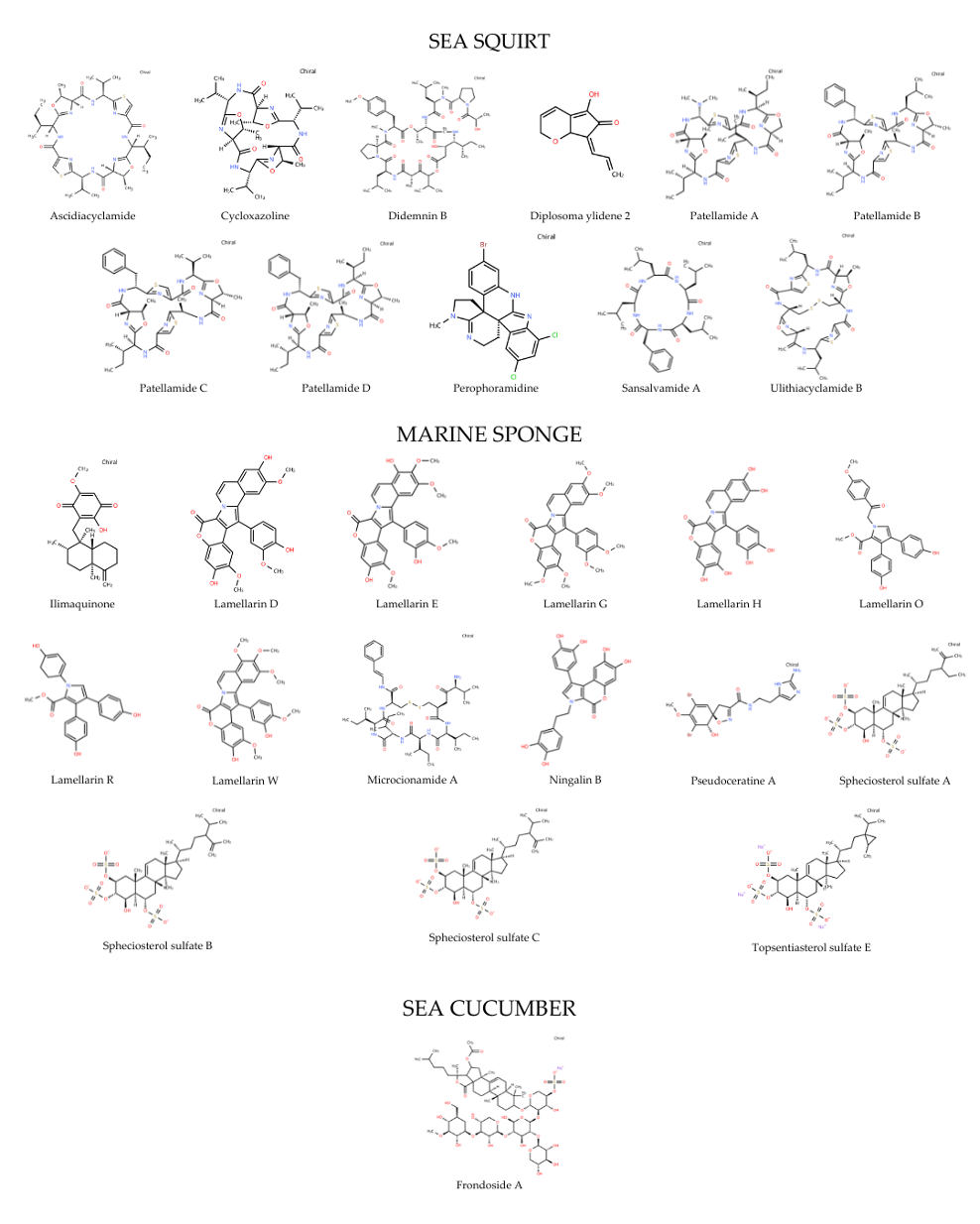
Since identifying effective compounds is time-consuming and costly, computational drug-discovery methods now enable high-throughput screening of bioactive molecules. Philippine marine natural products (MNPs) were computationally screened against four targets in the DNA damage response (DDR) pathways: Poly (ADP-ribose) polymerase 1 (PARP-1), Histone Deacetylase 2 (HDAC2), and topoisomerases I and II (TOPOI and TOPOII). These targets are crucial in various diseases, including cancer. A total of 27 MNPs were screened through molecular docking. Most favorable complexes were further analyzed for pharmacokinetic and drug-likeness profiling, visualization and molecular interaction analysis, and molecular dynamics (MD) simulations. Several MNPs demonstrated strong binding affinities to DDR targets, along with promising pharmacokinetic profiles supporting their drug candidacy including Ningalin B for PARP-1; Perophoramidine for HDAC2; Ulithiacyclamide B for TOPOI; and Patellamide C for TOPOII. MD simulations affirmed the stability of the best complexes via root-mean-square deviation and fluctuation, radius of gyration, and interaction energy analyses. Overall, the in silico results highlight the potential of Philippine MNPs—especially Ningalin B, Perophoramidine, Ulithiacyclamide B, and Patellamide C—as natural-product inhibitors of DDR pathways relevant to anticancer drug discovery.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering