VOLUME 19 NUMBER 1 (January to June 2026)
SciEnggJ. 2026 19 (1) 001-020
available online: 12 January 2026
DOI: https://doi.org/10.54645/2026191IQX-34
*Corresponding author
Email Address: aamanua@up.edu.ph
Date received: 06 October 2025
Dates revised: 26 November 2025
Date accepted: 02 January 2026
ARTICLE
Integrated bioinformatic analysis identifies key regulators and candidate biomarkers of cardiac fibrosis following acute myocardial infarction
Philippines, Diliman, Quezon City, 1101 Philippines
2National Research Council of the Philippines, Department of
Science and Technology, Bicutan, Taguig City, 1631 Philippines
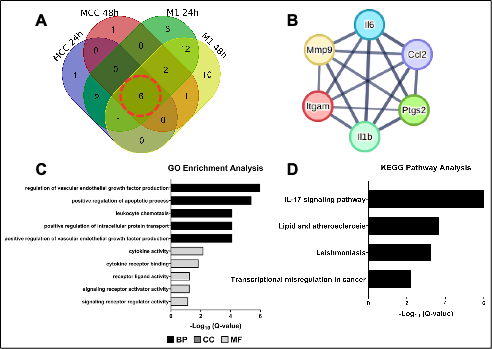
Cardiac fibrosis, a hallmark of numerous cardiovascular diseases, is strongly associated with adverse outcomes such as heart failure. However, current therapies remain ineffective in halting its progression, largely because the underlying mechanisms and pathways are poorly understood. Traditional laboratory validation of candidate biomarkers is often labor-intensive, costly, and time-consuming. Hence, computational approaches have emerged as powerful tools to accelerate the identification of candidate biomarkers. This study, therefore, aims to identify key regulators and candidate biomarkers associated with cardiac fibrosis following acute myocardial infarction (AMI). Using an integrated bioinformatic approach, we retrieved eligible microarray datasets from the Gene Expression Omnibus (GEO) repository, identify differentially expressed genes (DEGs), constructed protein–protein interaction (PPI) networks, identify hub genes, and predicts the transcription factor–gene regulatory relationships and microRNA–mRNA regulatory networks. As a result, we found only GSE775 and GSE4648 datasets that met our criteria. Across shared 24 h and 48 h time points, we identified 565 common DEGs. Functional enrichment analysis revealed that inflammatory response and IL-17 signaling pathway are the major contributors to cardiac fibrosis progression. Network-based analysis revealed six highly connected hub genes - Il1b, Itgam, Ccl2, Mmp9, Il6, and Ptgs2 - as central regulators of the fibrotic response. These hub genes are shown to be modulated by a network of transcription factors (NF-KB1, PPARA, FOS, EGR-1, and CEBPB) and microRNAs (mmu-miR-223-3p, mmu-miR-196b-5p, mmu-miR-181a-5p, mmu-miR-122-5p, and mmu-let-7c-5p), suggesting a multi-layered regulation of cardiac fibrosis progression. Collectively, the findings identify potential key regulators and candidate biomarkers of cardiac fibrosis following AMI. This integrative approach provides insights for future mechanistic studies, cross-species validation efforts and therapeutic exploration in cardiac fibrosis post-AMI.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering