VOLUME 19 NUMBER 1 (January to June 2026)
SciEnggJ. 2026 19 (1) 046-068
available online: 18 January 2026
DOI: https://doi.org/10.54645/2026191JVJ-47
*Corresponding author
Email Address: smdepaz@up.edu.ph
Date received: 13 June 2025
Dates revised: 14 November 2025, 23 December 2025
Date accepted: 30 December 2025
ARTICLE
IHC/FISH concordance and prognostic value of microarray-based genomic profiling tests in breast cancer molecular subtyping: A systematic review
2Virology Laboratory, Department of Medical Microbiology, College of Public Health, University of the Philippines Manila, Ermita, Manila, 1000 Philippines
3Department of Medicine, Philippine General Hospital, University of the Philippines Manila, Ermita, Manila, 1000 Philippines
4Lung Center of the Philippines, Diliman, Quezon City, 1100 Philippines
5College of Medicine, University of the Philippines Manila, Ermita, Manila, 1000 Philippines
6Department of Veterinary Paraclinical Sciences, College of Veterinary Medicine, University of the Philippines Los Baños, College, Los Baños, Laguna, 4031 Philippines
∆Co-First Authors
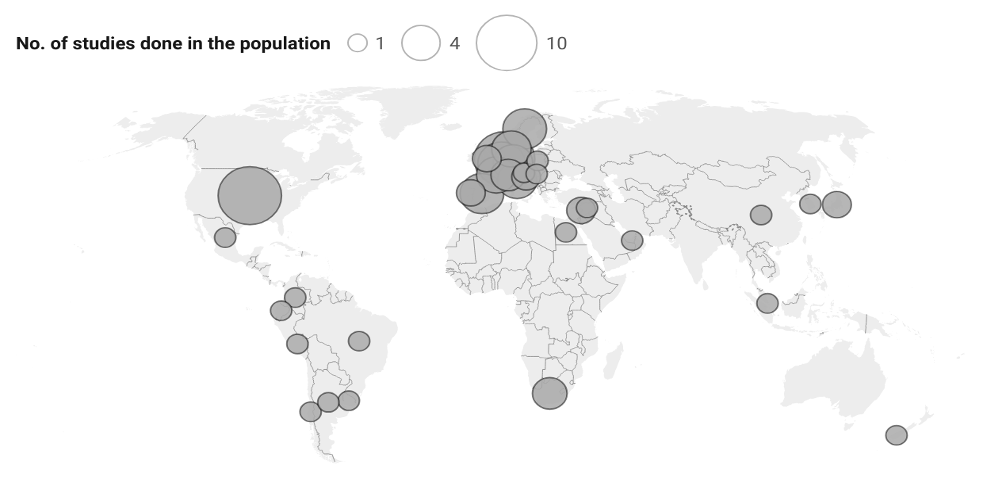
Breast cancer is a major public health concern worldwide. Accurate molecular subtyping is essential for appropriate patient management. Conventional pathology-based (immunohistochemistry/fluorescence in situ hybridization, IHC/FISH) tests have been the cornerstone of molecular subtyping, but newer genomic-based microarray techniques have shown promise to better classify tumors. Therefore, we aimed to systematically review the concordance and prognostic value of microarray-based breast cancer subtyping techniques. Scopus, Google Scholar, PubMed and Cochrane databases were systematically searched according to PRISMA guidelines. Articles comparing IHC/FISH- and microarray-based subtyping were included. Data on concordance, treatment response, and survival outcomes were extracted and analyzed. Independent reviewers assessed all articles against the inclusion and exclusion criteria. The concordance of microarray with IHC/FISH varied in the selected 45 articles. Studies using TargetPrint reported up to 100% concordance while one PAM50 study reported concordance rates as low as 54–60%. Some BluePrint-based microarray assays reclassified approximately 18% of luminal tumor to basal type, and 5% of ER+HER2- tumors to basal type. Microarray-based technologies have shown potential in prognosticating treatment outcomes and survival, and they may enhance breast cancer subtyping and improve the prediction of clinical outcomes. Future directions should focus on the standardization of microarray-based subtyping techniques and validating their clinical utility in various populations.
© 2026 SciEnggJ
Philippine-American Academy of Science and Engineering